-Search query
-Search result
Showing 1 - 50 of 294 items for (author: rein & a)

EMDB-19666: 
Cryo-electron tomogram of apoferritin
Method: electron tomography / : Comet M, Dijkman PM, Boer Iwema R, Franke T, Masiulis S, Schampers R, Raschdorf O, Grollios F, Pryor Jr EE, Drulyte I

EMDB-19667: 
Cryo-electron tomogram of keyhole limpet hemocyanin
Method: electron tomography / : Comet M, Dijkman PM, Boer Iwema R, Franke T, Masiulis S, Schampers R, Raschdorf O, Grollios F, Pryor Jr EE, Drulyte I

EMDB-19668: 
Cryo-electron tomogram of TGEV virions
Method: electron tomography / : Comet M, Dijkman PM, Boer Iwema R, Franke T, Masiulis S, Schampers R, Raschdorf O, Grollios F, Pryor Jr EE, Drulyte I

EMDB-19669: 
Cryo-electron tomogram of Influenza virions
Method: electron tomography / : Comet M, Dijkman PM, Boer Iwema R, Franke T, Masiulis S, Schampers R, Raschdorf O, Grollios F, Pryor Jr EE, Drulyte I

EMDB-19670: 
Cryo-electron tomogram of Magnetospirillum gryphiswaldense
Method: electron tomography / : Comet M, Dijkman PM, Boer Iwema R, Franke T, Masiulis S, Schampers R, Raschdorf O, Grollios F, Pryor Jr EE, Drulyte I

EMDB-19671: 
In situ cryo-electron tomogram of Saccharomyces cerevisiae
Method: electron tomography / : Comet M, Dijkman PM, Boer Iwema R, Franke T, Masiulis S, Schampers R, Raschdorf O, Grollios F, Pryor Jr EE, Drulyte I

EMDB-19672: 
Cryo-electron tomogram of keyhole limpet hemocyanin (stage position)
Method: electron tomography / : Comet M, Dijkman PM, Boer Iwema R, Franke T, Masiulis S, Schampers R, Raschdorf O, Grollios F, Pryor Jr EE, Drulyte I

EMDB-19673: 
Cryo-electron tomogram of keyhole limpet hemocyanin (3 um image shift)
Method: electron tomography / : Comet M, Dijkman PM, Boer Iwema R, Franke T, Masiulis S, Schampers R, Raschdorf O, Grollios F, Pryor Jr EE, Drulyte I

EMDB-19674: 
Cryo-electron tomogram of keyhole limpet hemocyanin (6 um image shift)
Method: electron tomography / : Comet M, Dijkman PM, Boer Iwema R, Franke T, Masiulis S, Schampers R, Raschdorf O, Grollios F, Pryor Jr EE, Drulyte I

EMDB-19064: 
CryoEM reconstruction of SARS-CoV-2 spike in complex with nanobody tri-TMH (partially open conformation)
Method: single particle / : Rissanen I, Hannula L, Huiskonen JT

EMDB-19068: 
CryoEM reconstruction of SARS-CoV-2 Spike in complex with nanobody tri-TMH (closed conformation)
Method: single particle / : Rissanen I, Hannula L, Huiskonen JT

EMDB-17350: 
Single particle cryo-EM co-structure of Klebsiella pneumoniae AcrB with the BDM91288 efflux pump inhibitor at 2.97 Angstrom resolution
Method: single particle / : Boernsen C, Mueller RT, Pos KM, Frangakis AS

PDB-8p1i: 
Single particle cryo-EM co-structure of Klebsiella pneumoniae AcrB with the BDM91288 efflux pump inhibitor at 2.97 Angstrom resolution
Method: single particle / : Boernsen C, Mueller RT, Pos KM, Frangakis AS

EMDB-18941: 
SARS-CoV-2 S (Spike) protein (BA.1) in complex with VHH Ma16B06 (sub-volume of two adjacent RBD-VHH modules)
Method: single particle / : Guttler T, Aksu M, Gorlich D

EMDB-18874: 
Cofactor-free Tau 4R2N isoform
Method: helical / : Limorenko G, Tatli M, Kolla R, Nazarov S, Weil MT, Schondorf DC, Geist D, Reinhardt P, Ehrnhoefer DE, Stahlberg H, Gasparini L, Lashuel HA

PDB-8r3t: 
Cofactor-free Tau 4R2N isoform
Method: helical / : Limorenko G, Tatli M, Kolla R, Nazarov S, Weil MT, Schondorf DC, Geist D, Reinhardt P, Ehrnhoefer DE, Stahlberg H, Gasparini L, Lashuel HA

EMDB-28036: 
Cryo-EM structure of the full-length human NF1 dimer
Method: single particle / : Darling JE, Merk A, Grisshammer R, Ognjenovic J

PDB-8edm: 
Cryo-EM structure of the full-length human NF1 dimer
Method: single particle / : Darling JE, Merk A, Grisshammer R, Ognjenovic J

EMDB-16452: 
Human spliceosomal PM5 C* complex
Method: single particle / : Dybkov O, Kastner B, Luehrmann R

PDB-8c6j: 
Human spliceosomal PM5 C* complex
Method: single particle / : Dybkov O, Kastner B, Luehrmann R

EMDB-27826: 
Cryo-EM structure of the full-length human NF1 dimer
Method: single particle / : Darling JE, Merk A, Grisshammer R, Ognjenovic J

PDB-8e20: 
Cryo-EM structure of the full-length human NF1 dimer
Method: single particle / : Darling JE, Merk A, Grisshammer R, Ognjenovic J

EMDB-16383: 
Cryo-EM map of SARS-CoV-2 S-trimer (3 RBDs up) in complex with neutralizing sherpabody TriSb92
Method: single particle / : Huiskonen JT, Rissanen I, Hannula L

EMDB-16388: 
CryoEM density of SARS-CoV-2 S spike (3 RBDs down)
Method: single particle / : Huiskonen JT, Rissanen I, Hannula L

PDB-8c1v: 
SARS-CoV-2 S-trimer (3 RBDs up) bound to TriSb92, fitted into cryo-EM map
Method: single particle / : Huiskonen JT, Rissanen I, Hannula L
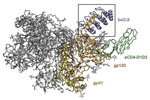
EMDB-26157: 
Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9
Method: single particle / : Cerutti G, Gorman J

PDB-7txd: 
Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9
Method: single particle / : Cerutti G, Gorman J, Kwong PD, Shapiro L

EMDB-26719: 
FMC63 scFv in complex with soluble CD19
Method: single particle / : Meyerson J, He C

EMDB-26720: 
SJ25C1 Fab in complex with soluble CD19
Method: single particle / : Meyerson J, He C

PDB-7urv: 
FMC63 scFv in complex with soluble CD19
Method: single particle / : Meyerson J, He C

PDB-7urx: 
SJ25C1 Fab in complex with soluble CD19
Method: single particle / : Meyerson J, He C

EMDB-25022: 
Cytoplasmic tail deleted HIV Env trimer in nanodisc
Method: single particle / : Yang S, Walz T
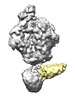
EMDB-25024: 
Cryo-EM map for HIV-1 Env bound with one 4E10 Fab
Method: single particle / : Yang S, Walz T

EMDB-25025: 
Cryo-EM map for HIV-1 Env bound with two 4E10 Fabs
Method: single particle / : Yang S, Walz T

EMDB-25045: 
Cytoplasmic tail deleted HIV-1 Env bound with three 4E10 Fabs
Method: single particle / : Yang S, Walz T

PDB-7sc5: 
Cytoplasmic tail deleted HIV Env trimer in nanodisc
Method: single particle / : Yang S, Walz T

PDB-7sd3: 
Cytoplasmic tail deleted HIV-1 Env bound with three 4E10 Fabs
Method: single particle / : Yang S, Walz T

EMDB-14049: 
Cryo-EM structure of ABC transporter STE6-2p from Pichia pastoris with Verapamil at 3.2 A resolution
Method: single particle / : Schleker ESM, Reinhart C

EMDB-14050: 
Cryo-EM structure of ABC transporter STE6-2p from Pichia pastoris in apo conformation at 3.1 A resolution
Method: single particle / : Schleker ESM, Reinhart C

PDB-7qkr: 
Cryo-EM structure of ABC transporter STE6-2p from Pichia pastoris with Verapamil at 3.2 A resolution
Method: single particle / : Schleker ESM, Reinhart C

PDB-7qks: 
Cryo-EM structure of ABC transporter STE6-2p from Pichia pastoris in apo conformation at 3.1 A resolution
Method: single particle / : Schleker ESM, Reinhart C
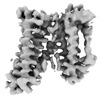
EMDB-24698: 
Cryo-EM structure of the HIV-1 restriction factor human SERINC3
Method: single particle / : Purdy MD, Leonhardt SA, Yeager M

EMDB-24705: 
Human SERINC3-DeltaICL4
Method: single particle / : Purdy MD, Leonhardt SA, Yeager M

PDB-7ru6: 
Cryo-EM structure of the HIV-1 restriction factor human SERINC3
Method: single particle / : Purdy MD, Leonhardt SA, Yeager M

PDB-7rug: 
Human SERINC3-DeltaICL4
Method: single particle / : Purdy MD, Leonhardt SA, Yeager M

EMDB-13680: 
Cryo-EM structure of small subunit of Giardia lamblia ribosome at 2.9 A resolution
Method: single particle / : Hiregange DG, Rivalta A, Bose T, Breiner-Goldstein E, Samiya S, Cimicata G, Kulakova L, Zimmerman E, Bashan A, Herzberg O, Yonath A

PDB-7pwf: 
Cryo-EM structure of small subunit of Giardia lamblia ribosome at 2.9 A resolution
Method: single particle / : Hiregange DG, Rivalta A, Bose T, Breiner-Goldstein E, Samiya S, Cimicata G, Kulakova L, Zimmerman E, Bashan A, Herzberg O, Yonath A

EMDB-13681: 
Cryo-EM structure of large subunit of Giardia lamblia ribosome at 2.7 A resolution
Method: single particle / : Hiregange DG, Rivalta A, Bose T, Breiner-Goldstein E, Samiya S, Cimicata G, Kulakova L, Zimmerman E, Bashan A, Herzberg O, Yonath A

EMDB-13683: 
Cryo-EM structure of Giardia lamblia ribosome at 2.75 A resolution
Method: single particle / : Hiregange DG, Rivalta A, Bose T, Breiner-Goldstein E, Samiya S, Cimicata G, Kulakova L, Zimmerman E, Bashan A, Herzberg O, Yonath A

PDB-7pwg: 
Cryo-EM structure of large subunit of Giardia lamblia ribosome at 2.7 A resolution
Method: single particle / : Hiregange DG, Rivalta A, Bose T, Breiner-Goldstein E, Samiya S, Cimicata G, Kulakova L, Zimmerman E, Bashan A, Herzberg O, Yonath A
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
